[root]/branches/henrik/ecoevo_ibm
![]() R
(4 files, 1126 lines)
R
(4 files, 1126 lines)
![]() output
(0 files, 0 lines)
output
(0 files, 0 lines)
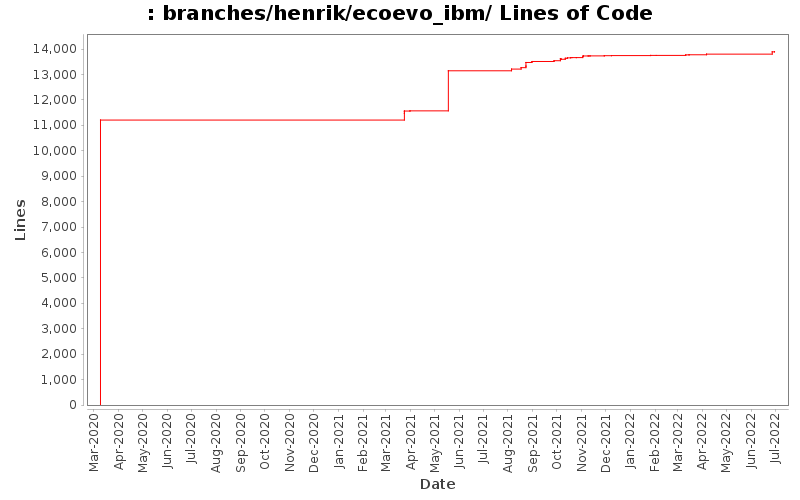
| Author | Changes | Lines of Code | Lines per Change |
|---|---|---|---|
| hje044 | 62 (100.0%) | 14954 (100.0%) | 241.1 |
Added functionality to saved the random seed in the ncdf file.
13 lines of code changed in 1 file:
Model version 1.0 finally ready. Now includes better functionality for starting from a previous population, plots have been added to showcase starting values, and year 1 is now always saved in results regardless of WriteInterval.
170 lines of code changed in 2 files:
Now also saves cumulative fecundity for plotting purposes.
45 lines of code changed in 1 file:
R script can now handle NA values, allowing me to load in runs that crashed prior to finishing
11 lines of code changed in 1 file:
Added option for increasing temperature over the years, and generalised parameters for stochastic deviance in inheritance and phenotypic plasticity.
80 lines of code changed in 1 file:
Includes better plotting functionality in R using interactive Plotly plots ans Esquisser() function built facet plots.
6 lines of code changed in 1 file:
Added diminishing returns to Appetite-energy, and moved Appetite onto energy intake rather than energy availability. Full rundown in OneNote 2022-2 and 2022-3.
44 lines of code changed in 1 file:
Moved density dependance to energy rather than appetite, testing out deminishing returns on appetite
14 lines of code changed in 1 file:
Makefile updated to be OS-agnostic. NetCDF libraries now pulled based on operating system
13 lines of code changed in 2 files:
Saved large runs after 10K years to reach equilibrium both fished and unfished
5 lines of code changed in 1 file:
New plots for wholly unfished population as well, use in meeting tomorrow
5 lines of code changed in 1 file:
Now plots evolution of maturation-related measures for all runs.
12 lines of code changed in 2 files:
Model now also capable of saving heritabilities. R-script updated to also pull in several runs heritabilities, and plotting these.
69 lines of code changed in 1 file:
Fixed a bug which added negative growth to smaller individuals, eventually leading to crashing the population
30 lines of code changed in 1 file:
Added biomass plot to R-script
1 lines of code changed in 1 file:
R loads parameters too, and now plots several runs alongside each other. Saved results include evolving traits, which R now plots over the years.
40 lines of code changed in 2 files:
Fixed bug in calculating skipspawnthreshold, number of spawn skippers now much more realistic, and no more negative gonad growth. Also added R functionality to read data when only a single run was made.
36 lines of code changed in 2 files:
R now capable of importing and combining multiple .nc files
1 lines of code changed in 1 file:
Now saves standard deviation, and avoids overwriting previous runs
95 lines of code changed in 2 files:
Cleaned up remnants of csv saving paradigm, and added legend to netCDF file. Also included new function to calculate standard deviation to be implemented in saving the results.
83 lines of code changed in 2 files:
netCDF file now also contains the parameters used in the saved run
58 lines of code changed in 1 file:
Added netCDF saving functionality to the fortran script, and rewrote R-script to load 3d netCDF data instead
97 lines of code changed in 3 files:
Improved efficiency in writing results, and added a makefile
58 lines of code changed in 3 files:
Model now saves files with years run in the filename to avoid overwriting when continuing population from earlier
94 lines of code changed in 1 file:
Added density dependant food environment, as well as saving results for these, and R plotting
36 lines of code changed in 2 files:
144 lines of code changed in 1 file:
Model now includes a recursive function for pulling random parents, which increase efficiency significantly. In addition, the model prints updates every few years, more stochasticity is included, and more comments have been added to increase readability.
125 lines of code changed in 1 file:
Added minimum energy for migrating, several types of stochastisticity, and most importantly: inheritance based on values from weighted random parent selection
80 lines of code changed in 1 file:
Added pop-size switch, included mixed fishing as GearType and tested random_normal() function.
104 lines of code changed in 3 files:
Added many comments for readability, and now scales recruitment to TargetInd (tentatively)
1653 lines of code changed in 2 files:
Fisheries mortality implemented. Calculated for Trawl, Gillnet and Unselective. R script now generates plots for population dynamics by year.
28 lines of code changed in 1 file:
Finally figured out how to properly set ignore for subfolders for all unversioned files.
16 lines of code changed in 2 files:
Attempting to clean up files and ignores
76 lines of code changed in 6 files:
Starting out using svn properly, hopefully
406 lines of code changed in 2 files:
Attempting to add the first version, which contains initialization, dummy growth, recruitment and age/removal
11206 lines of code changed in 6 files: